Wang, Y.; Liu, M.; Gao, J.. Proceedings of the National Academy of Sciences, 2020, 117 (25) 13967-13974
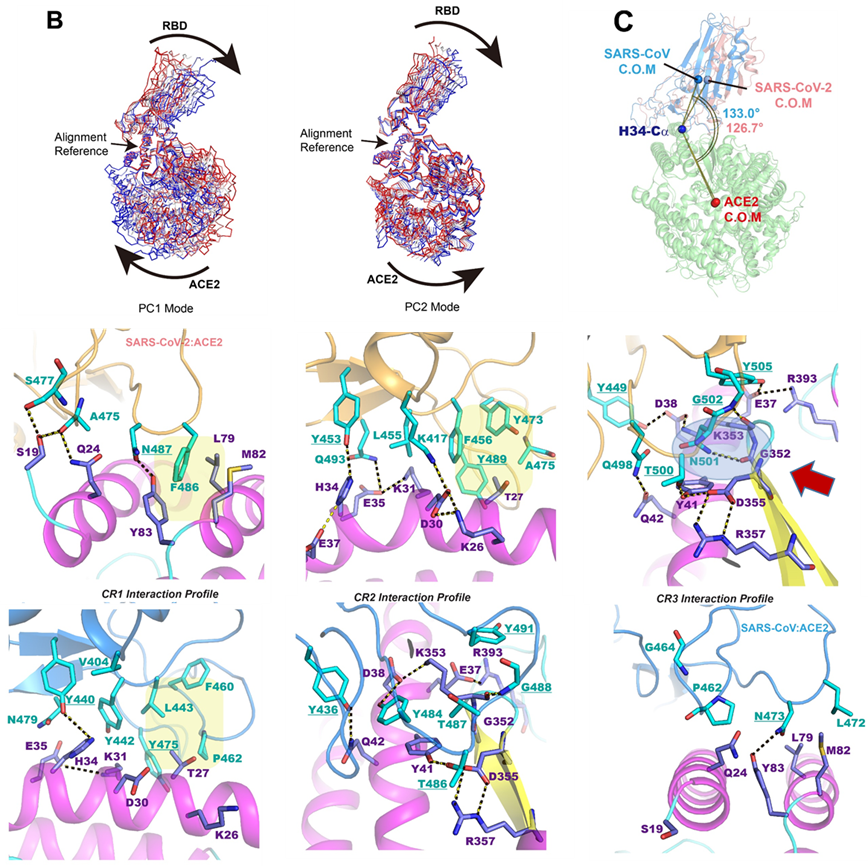
Molecular dynamics and free energy simulations have been carried out to elucidate the structural origin of differential protein–protein interactions between the common receptor protein angiotensin converting enzyme 2 (ACE2) and the receptor binding domains of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [A. E. Gorbalenya et al., Nat. Microbiol. 5, 536–544 (2020)] that causes coronavirus disease 2019 (COVID-19) [P. Zhou et al., Nature 579, 270–273 (2020)] and the SARS coronavirus in the 2002–2003 (SARS-CoV) [T. Kuiken et al., Lancet 362, 263–270 (2003)] outbreak. Analysis of the dynamic trajectories reveals that the binding interface consists of a primarily hydrophobic region and a delicate hydrogen-bonding network in the 2019 novel coronavirus. A key mutation from a hydrophobic residue in the SARS-CoV sequence to Lys417 in SARS-CoV-2 creates a salt bridge across the central hydrophobic contact region, which along with polar residue mutations results in greater electrostatic complementarity than that of the SARS-CoV complex. Furthermore, both electrostatic effects and enhanced hydrophobic packing due to removal of four out of five proline residues in a short 12-residue loop lead to conformation shift toward a more tilted binding groove in the complex in comparison with the SARS-CoV complex. On the other hand, hydrophobic contacts in the complex of the SARS-CoV–neutralizing antibody 80R are disrupted in the SARS-CoV-2 homology complex model, which is attributed to failure of recognition of SARS-CoV-2 by 80R.